Hierarchy¶
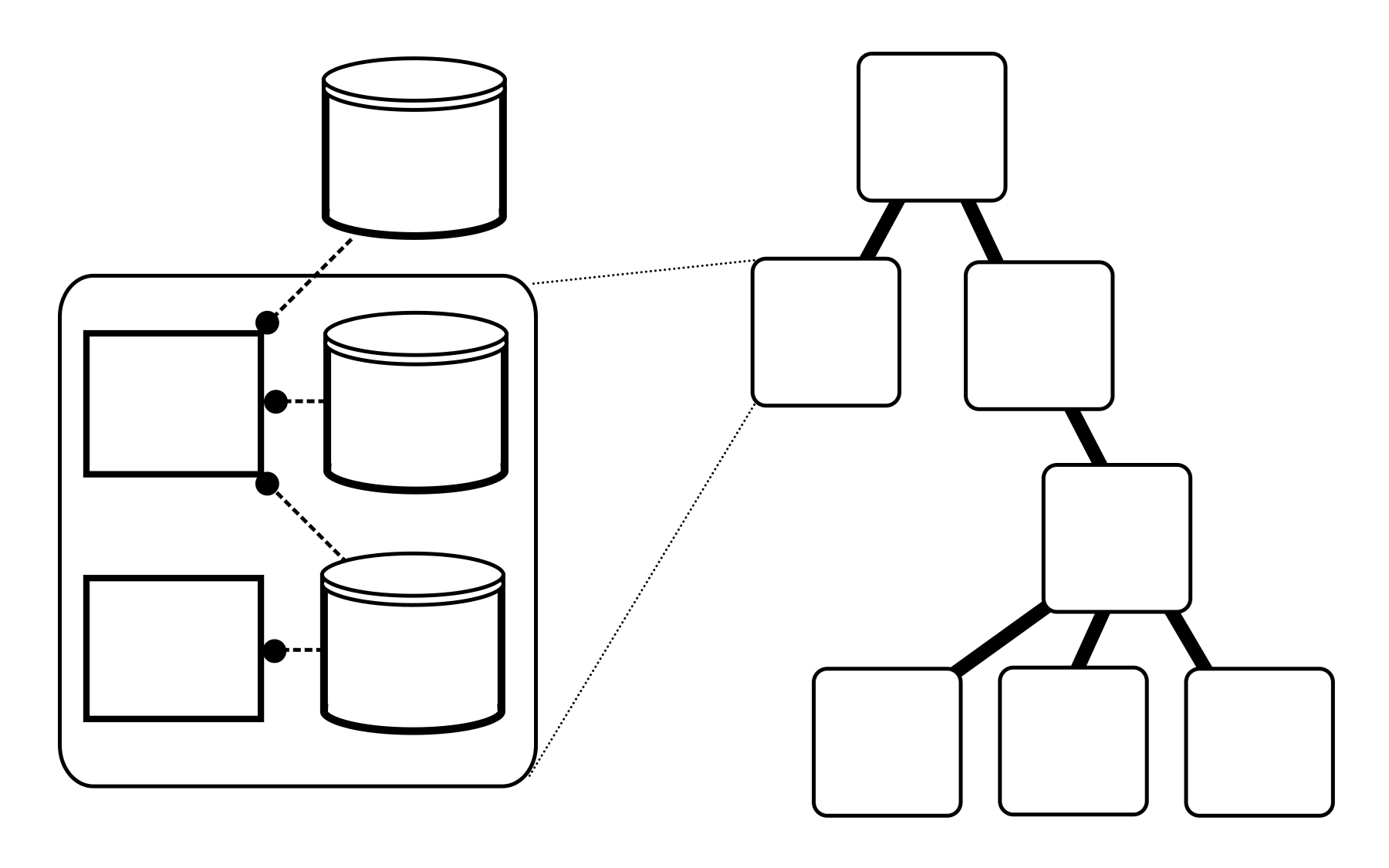
In Vivarium, we represent the simulation as a tree of processes and stores (panel A). The processes and stores are wired together by topologies to form compartments (panel B), which can then be nested to form a tree called the hierarchy (panel C).

The relationships between stores, processes (panel A), and compartments (panel B) in the hierarchy (panel C).¶
Note that in panel C, only the compartments and boundary stores are shown. The full hierarchy also contains the stores and processes within each compartment.
Note
Here we have shown a simplified picture where processes are only wired to stores in their own compartment or to boundary stores. In reality, processes can be wired to any store in the hierarchy, but keeping cross-compartment wiring to a minimum can help simplify your models.
We recommend using Vivarium compartments to represent spatial regions of your modeled system that are conceptually distinct. For example, you might model a cell as a compartment and its environment as another compartment. This is not a technical requirement though, so you can use compartments to represent whatever makes sense for your circumstances.
Compartment Interactions¶
We model cross-compartment interactions using boundary stores between compartments. For example, the boundary store between a cell and its environment might track the flux of metabolites between the cell and environment compartments.
When compartments are nested, these boundary stores also exist between the inner and the outer compartment. Thus nested compartments form a tree whose nodes are compartments and whose edges are boundary stores. A node’s parent is its outer compartment, while its children are the compartments within it.
Since boundary stores can also exist between compartments who share a parent, you may find it useful to think of compartments and their boundary stores as a bigraph (not a bipartite graph) where the tree denotes nesting and all the edges (including those in the tree) represent boundary stores.
Hierarchy Structure¶
In the example below, we print out the full hierarchy as a dictionary.
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.engine import Engine
>>> from vivarium.core.composer import Composer
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
Notice that in the dictionary above, each leaf node in the tree is a key with a value that is a dictionary of schema keys.
Hierarchy Paths¶
A hierarchy in Vivarium is like a directory tree on a filesystem. In
line with this analogy, we specify nodes in the hierarchy with paths.
Each path is a tuple of node names (variable names or store names)
relative to some other node. For example, in the topology from the
example above, we used the path ('cell', ) to say that the cell
store maps to the injector’s internal port. This path was
relative to the compartment root (root in our diagram) as is the
case for all topologies. Thus the path is analogous to ./cell in a
directory.
Special Symbols¶
Continuing our analogy between hierarchy paths and file paths, the following symbols have special meanings in hierarchy paths:
..refers to a parent node. One example use for this is a division process that needs to access the parent (environment) compartment to create the daughter cells. In fact, this is what we do in the growth and division compartment.